A Deep Learning Chatbot Simulation of Clinician Conversations

Intro
The goal of this project is to showcase a proof-of-concept for generating appropriate responses to health-related questions. While the medium of interaction, such as online, email, or face-to-face, would impact the approach for any full-scale prototype, having a minimal viable product to showcase the approach is essential.
As a bootstrapped initiative, the focus is on sharing safe content with the audience without exposing any patient data. The envisioned AI system would interact with the user in a way that complements a doctor’s expertise, but never replacing the final say. However, this is a highly complex problem, and reducing it to a simpler problem is a crucial step. For example, given a statement or query about someone’s health, what would be an appropriate response from a clinician?
This problem is only one step towards creating a comprehensive system that can integrate with platforms such as Google voice or Alexa, once they determine that the user is discussing their health. The aim of this project is to lay the foundation for such a system, exploring the possibilities of generating suitable responses to health-related inquiries. A minimum viable product will serve as a proof-of-concept and demonstrate the approach, with the potential to revolutionize the healthcare industry by providing accessible and accurate responses to health-related queries in real-time.
General approaches to machine generated conversational responses
Although it seems like most developed solutions are a little mix-and-match, these categories are generally how most people would bucket at chatbot approach. Researching these was particularly helpful in characterizing my understanding.
-
Rule-base: This is along the lines of: given a serious question about heart attack. Tell them to call 911. Not robust. Hard to maintain. Etc…
-
Selective: There are many different approaches that can be used for selecting the best response from a bank of potential responses, but one of the most robust and time-tested approaches is using a classifier. Essentially, a classifier is used to determine the best response(s) based on certain criteria, such as relevance or accuracy. This approach has been widely used in various fields, including natural language processing and information retrieval.
-
Generative: Another approach that has been explored in natural language processing is autoencoding, also known as autoencoders. This technique involves predicting the next sequence of characters or tokens given a sequence of input tokens. However, autoencoding may not be the best choice for this project due to its tendency towards randomness, especially in a medical context where the vocabulary can be complex and specialized. While significant advances have been made in this area, sparsely used words in the medical domain can still pose a challenge for autoencoding-based models.
While a rule-based and generative approach may have appeared suitable at first, it is worth noting that these models can be limited in their capacity to capture the nuances of natural language and human conversation. Specifically, a rule-based approach may struggle with the complex and diverse language used in clinical settings, which can include medical jargon, colloquialisms, and other domain-specific language.
Furthermore, while offering clinicians control over the response selection process may appear advantageous, it can also introduce bias into the system and restrict the variety of responses generated. This is especially critical in the healthcare field, where a wide range of perspectives and approaches may be required to provide effective care to patients. As a result, a more adaptable and data-driven approach that can learn from a diverse range of inputs may be more appropriate for generating responses to health-related queries.
This led to the well-known paper The Ubuntu Dialogue Corpus: A Large Dataset for Research in Unstructured Multi-Turn Dialogue Systems (here) which introduces Ubuntu data to model appropriate responses to technical questions on a public forum. Coincidentally, the dataset that I had chosen was also based on public forum data, Reddit’s AskDocs Forum (here).
In the context of clinical conversations, a seq-to-seq model can be trained to map a sequence of input utterances from the patient to a sequence of output utterances from the clinician. This type of model can capture the context and flow of the conversation, as well as the nuances of language and domain-specific terminology that are unique to clinical settings.
Furthermore, a seq-to-seq model can incorporate attention mechanisms that allow the model to focus on specific parts of the input sequence when generating the output sequence. This can be particularly useful in clinical conversations, where certain aspects of the patient’s history or symptoms may be more important than others.
Dataset Description
It took some exploring, but I found a suitable, health-centric, conversational dataset in Reddit. There is a sub-forum (“subreddit”) called “AskDocs” where certified health professionals reply to questions by other forum members. The health professionals could be anyone from health students, nurses, to residents, to full-scale general practitioners, but had to provide evidence of their status to reddit moderators and can be identified as such. The data is updated and stored in Google’s BigQuery, however if you wish to download the data I used you can do directly at this link.
Formatting data and cleaning
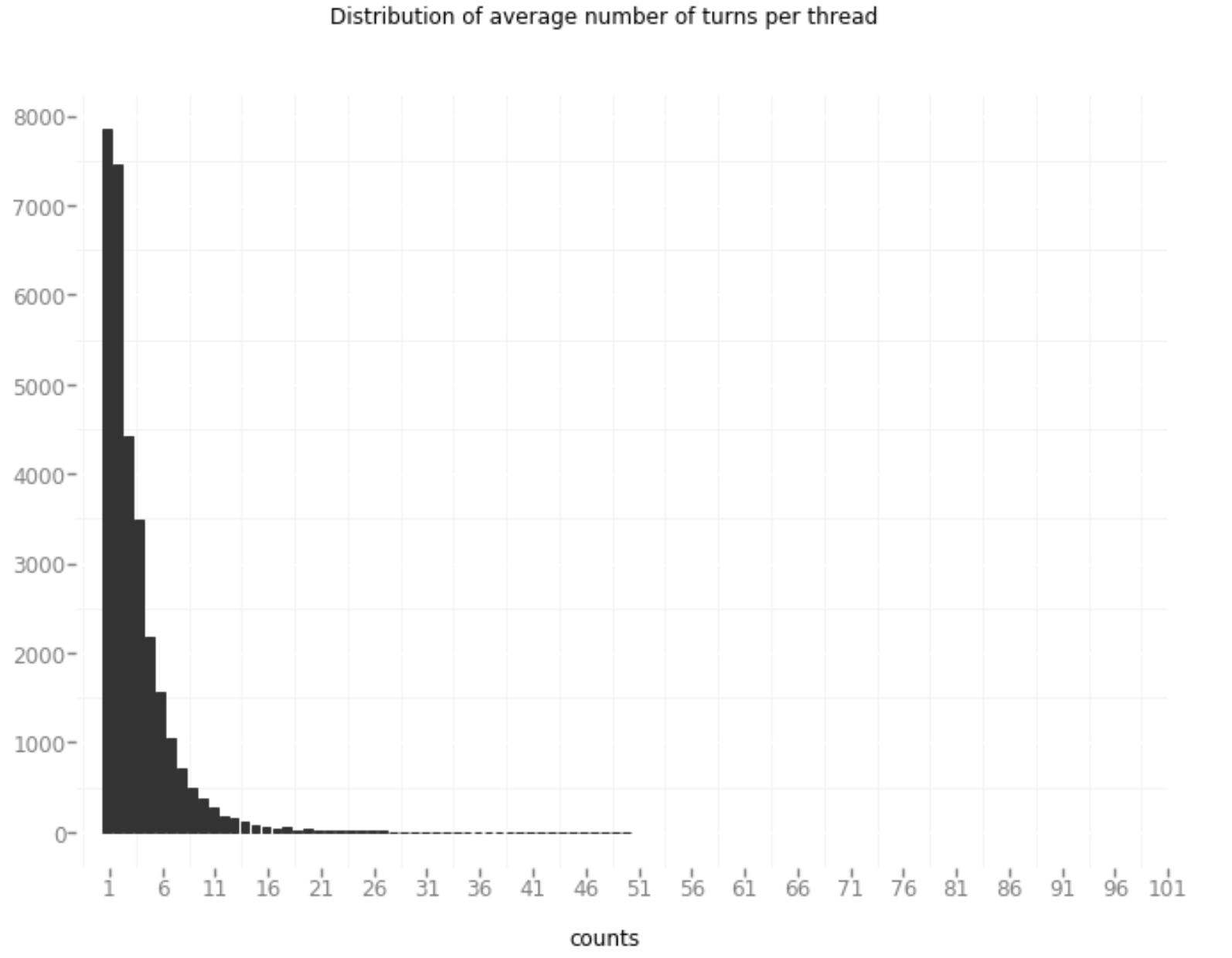
In order to model forum data the form of a query and response, the authors of the Ubuntu paper had to estimate (using a time window) whether a new post was in direct response to a previous post. With the Reddit data, the same approach certainly could have been applied. However, in Reddit, it is explicit as to whether a new post under a thread (an ongoing conversation concerning a topic started by the original post) is in direct response to the original post or in response to another comment. Also, per the paper’s recommendation, only threads with at least 3 posts were retained in order to provide context. The lower-cased form of all text was used along with standard tokenization using Keras. In total this generated 955,610 samples of unique 282,433 words which was split into train/test/validate 80/10/10 respectively:
A key way of measuring how dynamic a conversation is for a thread is counting the number of turns in a conversation (or thread). Turns refer to the switching between speakers. This is follows the assumption for human-like chatbots that conversation partners take turns in a conversation (or thread). Given that most of the posts in AskDocs only generate around 4-5 posts it is a safe assumption to say that all posts following that initial post are generally on the same topic. (great Medium post here on humanizing your chatbot)
The Model
To briefly summarize what we’re modeling: For a given query (i.e. question), can we make a binary selection from a list of possible answers saying if the answer is appropriate.
I utilized code from a blog post (Implementation of dual encoder using Keras) on the implementation of dual encoders using Keras, making some updates to suit my specific needs. I highly recommend reading the blog post and the paper it’s based on for a more comprehensive understanding of the model. In testing, both the 6B token and 840B token Glove embeddings were utilized, with substantial improvements seen when using the 840B tokens. Additionally, embeddings trained on all 2015 Reddit comments were tested but showed no significant decrease in loss.
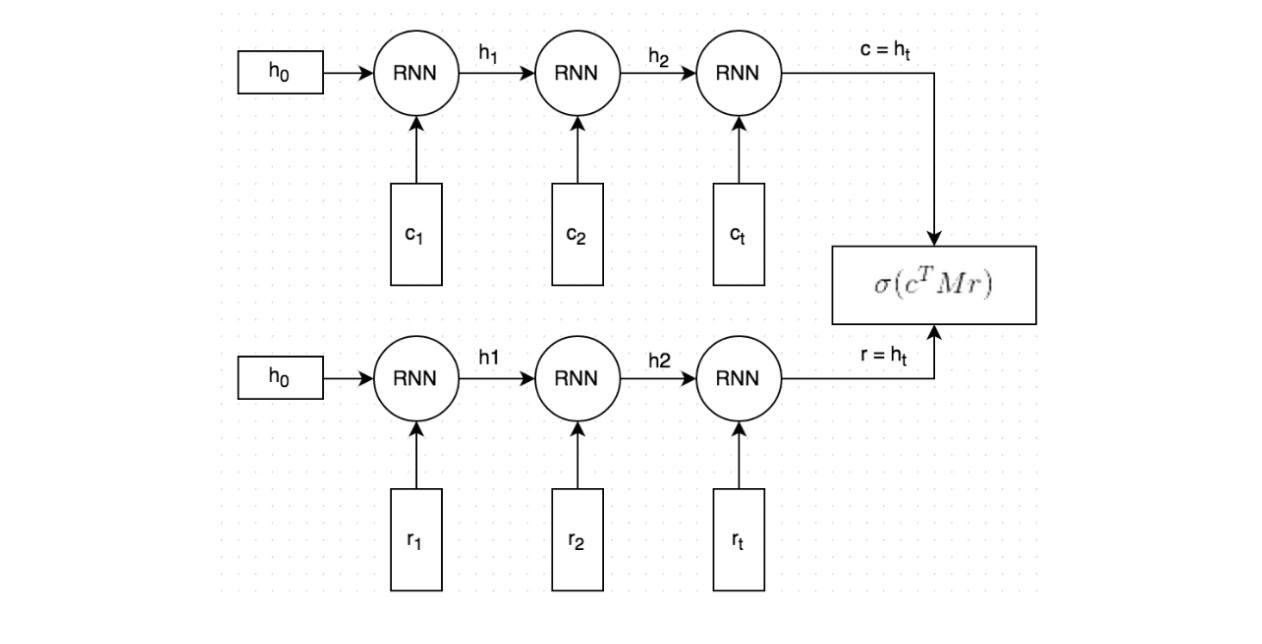
The main structural advantage of this model is that information is mapped in pairs. This is achieved in Keras by defining a sequential model with an embedding layer initialized with GLoVe embeddings. To enhance the performance of the model, I allowed these embeddings to be fine-tuned during the training process. By doing so, the model can learn to represent the data more accurately and improve its ability to make predictions.
Results
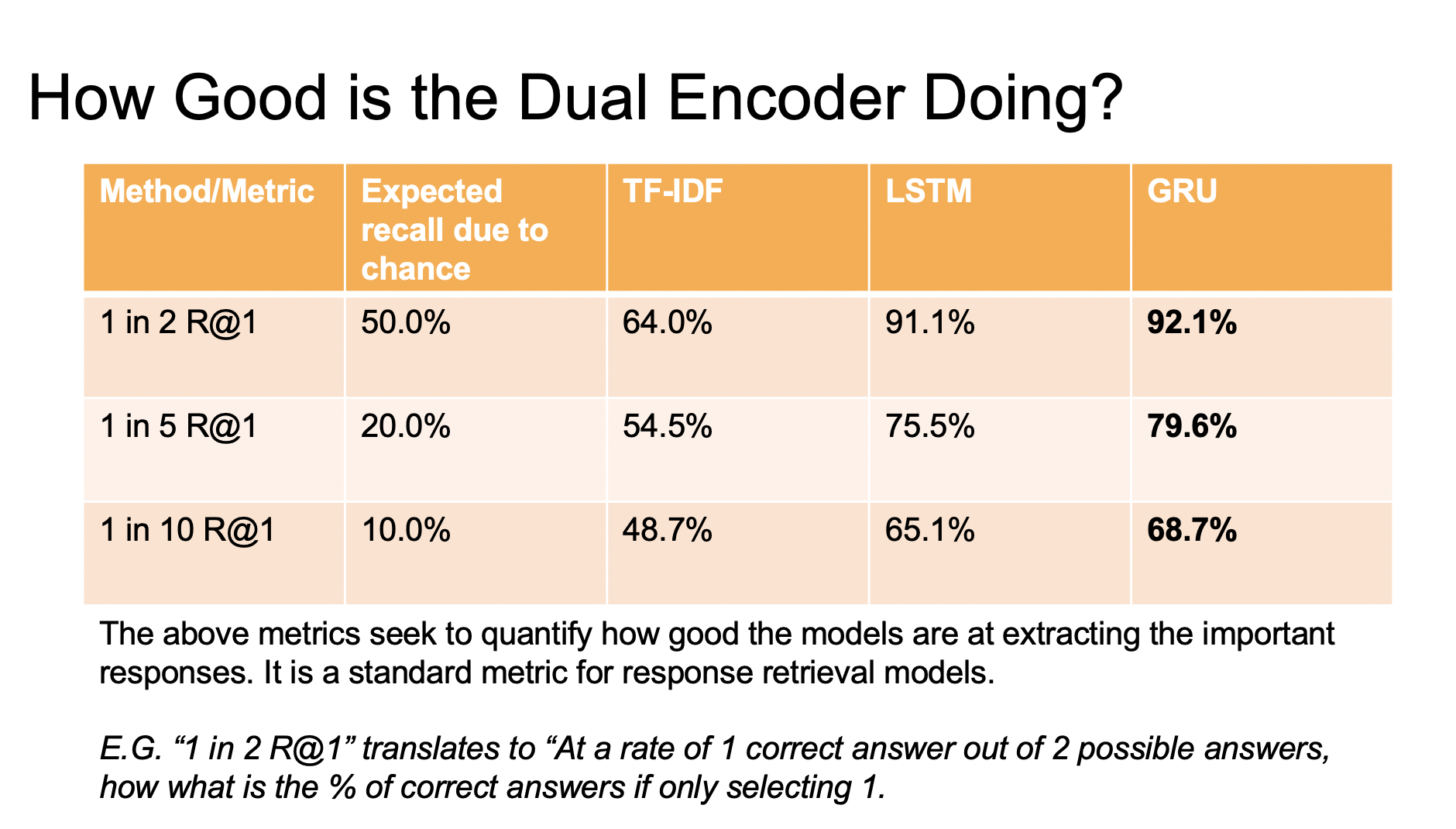
Conclusion
The objective of this project was to adapt the Ubuntu Dual Encoder paper for health data. Comparing base metrics to the original paper showed similar results, with minimal model tuning and data parsing adjustments. However, unlike the original paper, comments prior to a given query were omitted to provide context in selecting responses. Despite this difference, results from the health data are promising and indicate potential for creating more satisfying and health-centric responses. If successful, this technology could help the healthcare industry by providing accessible and accurate real-time responses to health-related queries. It’s important to note that the quality of the dataset used plays a significant role in the effectiveness of the approach, and further research is necessary to test its generalizability on other datasets.
Papers
- Diagnosing Disease: A Self-Diagnosis Medical Chatbot Using Artificial Intelligence
- Helped to decide on model for retrieval: The Ubuntu Dialogue Corpus: A Large Dataset for Research in Unstructured Multi-Turn Dialogue Systems
Dataset creation
- There are a few changes to how the data was manipulated that might train a model for an appropriate response. For example the model currently is train on a response to every query so there can be multiple responses to the same query. Using something like the number of upvotes, one response could be selected (at least for the initial query).
- Conversation history in the thread could be include in the encoder portion of the model. This would potentially help the correct results so that we would be able to select the most relevant response given the history as opposed to a technically correct, but non-sensical result like the one below.

Refinement of “Health” purposes
Most of this work was towards creating an initial viable solution in human-like responses to a health-related statement or question. One way of refining the types of responses would be to determine what type of statement/questions started the conversation and score possible answers only within that concept space.
Code for this project can be viewed at: Automated Health Responses